Animal Influenza Ecology and Epidemiology Research Program
The College of Veterinary Medicine's Animal Influenza Ecology and Epidemiology Research Program is dedicated to the advancement of knowledge and the safeguarding global health. At the heart of our initiative lies a profound vision: to prevent zoonotic disease outbreaks, epidemics, and pandemics across all species, including humans. Our mission is to serve as a collaborative leader at the forefront of viral zoonoses, shaping the interface between animals and humans, and developing innovative preventive strategies to fortify both animal and public health.

Our Goals
- Reduce viral sources associated with the emergence of novel zoonotic diseases through surveillance and ecology and epidemiology research efforts in: (1) wildlife across the Midwest; and (2) the swine-human interface in commercial settings and agricultural fairs/exhibitions.
- Identify risk factors and criteria for intra- and inter-species transmission.
- Utilize virus recovery, viral genomics, and host ecology to better understand the natural history of type A viruses.
- Educate the public about the spread of disease, how to limit the transmission and biosecurity protocols.
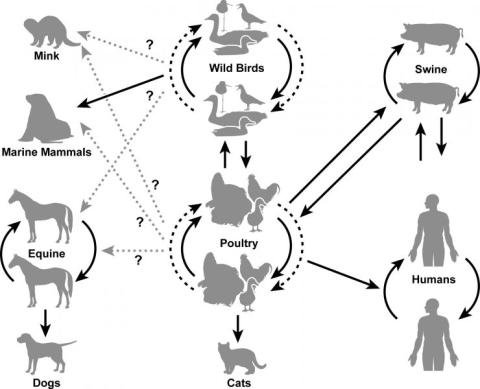
Background Information on Influenza in Animals
Influenza viruses are RNA viruses in the family Orthomyxoviridae. The influenza viruses are categorized into three main types based on their antigenic properties: A, B, and C.
- Type A influenza viruses are the most virulent and are maintained in humans, lower mammals, and birds.
- Type B and C influenza viruses are only maintained in humans.
Influenza A viruses are divided into subtypes based on two glycoproteins on the surface of the virus:
- Hemagglutinin (HA): 18 different subtypes
- Neuraminidase (NA): 11 different subtypes
Influenza A subtype combinations are further classified by strains (using genetic sequencing).
- Avian influenza virus strains are further classified as either low pathogenic (LPAI) or highly pathogenic (HPAI) based on specific molecular and pathogenesis criteria that require additional specific testing.
- Classification as low or high pathogenicity (severity of disease) is specific to poultry, and not necessarily to other animal species that can be susceptible to avian influenza viruses including humans.
- Only H5 and H7 subtypes have shown the capability of being HPAI.
- Antigenic drifts can result in minor antigenic changes in the HA and NA creating a new strain (same subtypes).
- Antigenic shifts can result in a sudden appearance of a new strain possessing a distinctly different HA and/or NA subtype.
Influenza type/host of origin/geographic source/isolate number/year of isolation (HA subtype NA subtype)
- Example: Influenza A/long-tailed duck/WI/10OS3912/2010 (H14N6)
- Humans: H1N1, H2N2, H3N2; Since 1997: H5N1, H7N2, H7N3, H7N7, H7N9, H9N2
- Swine: H1N1, H3N2, H1N2; (H3N1, H4N6, H5N1, H9N2)
- Equine: H7N7, H3N8
- Poultry: Many HA NA combinations recovered; not all HA NA subtypes recovered.
- Wild birds: Many more HA NA combinations recovered; all HA NA subtypes recovered (except newly discovered subtypes which have only been found in bats).
- Wild waterfowl are the primary natural reservoir for all subtypes of influenza A viruses and are believed to be the original source of influenza A viruses in all other animals.
- In wild birds, most influenza viruses cause asymptomatic (subclinical) or mild infection; however, the range of symptoms varies greatly depending on the strain of virus.
- Influenza viruses typically infect the avian gastrointestinal tract rather than the respiratory tract; thus, virus is shed through feces.
SARS-CoV-2 is an emerging virus that first appeared in late 2019 in humans. During the 2020-2021 hunting season, the first positive cases of SARS in white-tailed deer appeared in Northeast Ohio and has since been found across 24 states. In Ohio, samples have been taken from 83 of 88 counties and of those 83 counties, 50 have had at least one positive deer sample.
Not much is yet known about infection in deer, include symptoms and transmission from humans to deer, deer to deer and other species to/from deer. Possible theories of transmission from humans to the deer include backyard compost piles, baits, and other human waste that could be contaminated with the virus, however, a conclusive answer has not been found. Deer are social animals, living in groups, making it easy to spread the virus through physical touch or food sharing. Currently, there is no evidence that white-tailed deer are reservoir hosts for deer to human transmission.
The purpose of our research and surveillance is to collect data to help understand the spread and transmission of this emerging virus in other species. With little known, it is important to collect and survey data to help regulatory officials make evidence-based decisions.
Read more about it here:
Wild waterfowl are recognized as the major reservoir for influenza A viruses which can infect poultry, swine, and humans. Waterfowl often show no clinical signs when infected with the virus making them a good source for transmission. Influenza A is an enteric virus in waterfowl, which means that it infects their digestive system, in contrast to the respiratory infection in humans and other animals. This enteric infection of waterfowl causes the virus to be passed in their feces, allowing for wide spread transmission within the waterfowl population and to other species during the annual bird migration.
Commercial and domestic poultry are particularly susceptible to the viruses passed by waterfowl. In chickens, influenza A viruses are classified as Low Pathogenic Avian Influenza (LPAI) and High Pathogenic Avian Influenza (HPAI). LPAI viruses tend to cause only minor illness and little to no mortality in chickens while HPAI viruses cause major illness and high mortality. Both LPAI and HPAI present major threats to the commercial poultry industry. Though LPAI present little immediate threat, influenza A viruses can rapidly mutate and LPAI viruses could mutate to HPAI viruses, leading to major commercial losses. Infection in a commercial flock often leads to large amounts of culling.
In 2022, there is an ongoing outbreak of HPAI H5N1. Positive cases have been reported in at least 34 states and 33 EU/EAA countries. USDA data reports, in the US, approximately 1200 wild birds, 141 backyard flocks and 178 commercial flocks have tested positive through extensive surveillance efforts. The number of birds affected has reached 37.87 million.
Influenza A infection of waterfowl and chickens also presents an infection risk to humans. According to the CDC, as of early May 2022, there has only been one positive human case of H5N1 in the US, however, the risk for the general public is low. Those who work with birds, are at higher risk of getting infected.
Another example of avian-origin influenza infecting people is the LPAI H7N9 endemic that occurred in China. With thriving live bird markets, chickens infected with the LPAI H7N9 virus and showing no signs of infection were transmitting the virus to humans handling them. According to the CDC, 40% of those infected have died and, even though there has been limited evidence of human-to-human transmission, H7N9 presented a risk for a global pandemic.
In order to keep an eye on the influenza A viruses circulating the waterfowl in the United States, our program does yearly surveillance during the annual bird migration. With the help of collaborators, wildlife biologists, and numerous hunters, we are able to collect thousands of samples from waterfowl each migration.
Swine are a source of novel and existing influenza A virus strains that infect humans. Interspecies transmission of viruses is a principal mechanism contributing to the emergence of novel influenza A virus strains that pose a threat to human and swine health. Pig respiratory tracts have receptors for swine-, human-, and avian-origin influenza A virus, which facilitates genomic reassortment among viruses from multiple host species. As a result, swine have been identified as mixing vessels for influenza A viruses and a source of emergence for novel viruses.
These virus strains pose a pandemic threat if they become capable of being transmitted efficiently from person to person and if limited protective immunity exists in the human population. Bidirectional interspecies transmission of influenza A virus strains usually involves close contact between humans and swine. The United States has 3 major swine–human interfaces: commercial swine production, abattoirs (slaughterhouses), and agricultural fairs.
Agricultural fairs/exhibitions are unique and important because they facilitate prolonged commingling of pigs from numerous sources raised under varied management programs with millions of persons who have widely disparate histories of exposure to various influenza viruses. This situation creates an environment conducive to zoonotic transmission of influenza A viruses. More persons come in contact with live swine at agricultural fairs than in any other setting in the United States, and the majority of human cases of swine-origin influenza A have been linked to swine exposure occurring at fairs.
The purpose of our influenza research and surveillance efforts with exhibition swine is to proactively collect scientific data to help regulatory officials make evidence-based decisions. In addition, we are actively evaluating preventive strategies to protect animal and public health. It is important to remember that influenza viruses are not transmitted by eating properly prepared pork and pork products.
- Conduct surveillance for influenza A virus circulating among exhibition swine at agricultural fairs and exhibitions.
- Conduct diagnostic testing of each sample for the presence of influenza A virus, including virus isolation and RRT-PCR, and maintain a repository of swine-origin isolates.
- Determine the HA and NA subtypes and submit isolates for full length genomic sequencing and subsequent placement into GenBank and IRD for use by scientists around the world.
- Evaluate genomic segments to determine differences in the genomic constellations of the recovered viruses.
- Look at possible swine-human transmission associated with agricultural exhibitions.
Take a look at some of our swine surveillance data below!
- Listen to Dr. Bowman's Podcast
- Measures to Minimize Influenza Transmission at Swine Exhibitions
- National Pork Board
Link to YouTube: https://www.youtube.com/watch?v=UhnGak-SrvI&embeds_euri=https%3A%2F%2Fv…
AIEERP in the News
- https://www.tampabay.com/news/environment/2022/04/26/avian-influenza-outbreak-has-spread-to-tampa-bay/
- https://www.nature.com/articles/d41586-022-01112-4
- https://health.osu.edu/community-health/health-and-society/the-us-has-one-of-the-deadliest-bird-flu-outbreaks
- https://www.nytimes.com/article/bird-flu.html
- https://www.nifa.usda.gov/about-nifa/blogs/womens-history-month-spotlight-dr-jacqueline-nolting
- https://abcnews.go.com/US/scientists-find-1st-deer-infected-omicron-variant-york/story?id=82744120
- https://thehill.com/changing-america/well-being/588238-scientists-say-covid-19-outbreak-in-deer-could-mean-trouble-for/
- https://www.cidrap.umn.edu/?f%5B0%5D=field_related_topics%3A71





