Sanggu Kim
Associate Professor

Contact
kim.6477@osu.edu (614) 292-6524 Veterinary Medicine Academic Building1900 Coffey Road
Columbus, OH 43210
Map Link
Department
Veterinary Biosciences
Center For Retrovirus Research
Professional Training and Experience
- Associate Professor, The Ohio State University
- Assistant Professor, The Ohio State University
- Assistant Researcher, UCLA MIMG
- Postdoctoral Training: UCLA AIDS Institute
- PhD: UCLA Biomedical Engineering
Research Overview
Research in the Kim laboratory focuses on systems-level analysis of the immunological processes and pathogen evolution that occurs in cell and gene therapy settings, particularly HIV/AIDS and cancer treatment. We develop novel genomics and systems-biology tools and employ conventional as well as cutting-edge research tools to better understand the interactions between host systems and pathogens and the molecular and cellular mechanisms underlying these events.
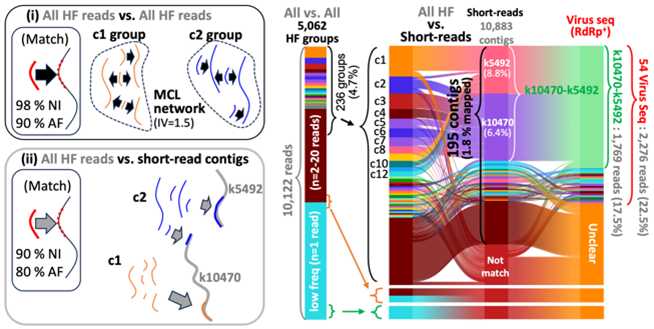
Read-level viral variant detection and environmental RNA virus discovery using high-fidelity Nanopore sequencing
Yu et al. 2025, Advanced Science
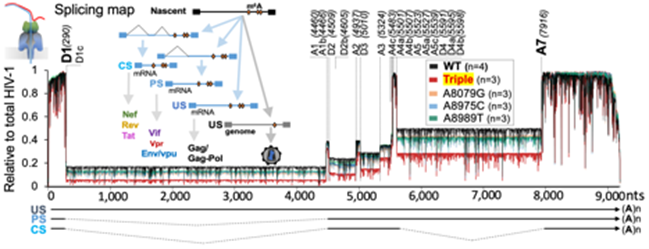
Single-RNA and nucleotide-resolution analysis of m6A modifications on HIV-1 RNA using Nanopore sequencing and oligonucleotide LC-MS/MS.
- Baek et al. 2024, Nature Microbiology (Accepted)
- Baek et al. 2024, Methods Protoc
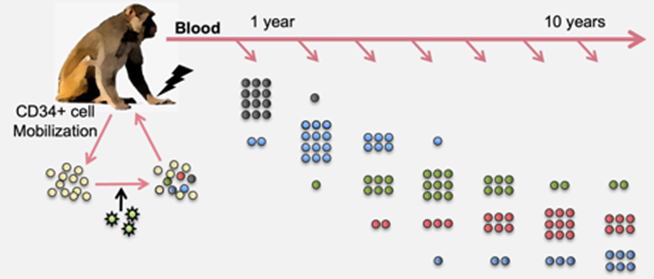
HIV-1 pathogenesis and cellular dynamics analysis at the single-cell and single-virus level in animal models of HIV-1 gene therapy.
- Gajendra et al., 2020, Science Advances
- Kim et al., 2014, Cell Stem Cell
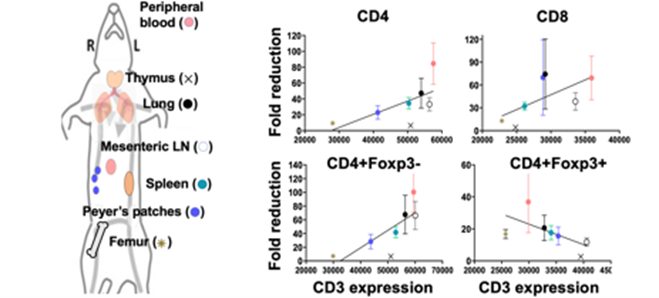
Systems-level, multi-organ evaluation of T-cell dynamics after T-cell depletion.
- Kim et al., 2022, Frontiers in Immunology
- Kim et al., 2022, Biomedicines; Wang et al., 2023, Heliyon
Research Interests
- Clonal HSPC and T-cell dynamics in HIV-1 gene therapy
- Virus-host interactions
- HIV-1 RNA post-transcriptional modifications
Current Research Support
- 2024 - 2026 Center for RNA Biology (co-PI: Kim) "Post-transcriptional modifications in HTLV-1 and ATLL development"
- 2024 - 2026 Canine Grant (PI: Kim) ”Developing an Advanced, Safety-Enhanced Immunotoxin Targeting Canine T Cells"
- 2023 - 2026 Department of Defense (PI: Kim) "Targeted antibody and immunotoxin combination improve T-cell lymphoma therapy"
Memberships in Professional Societies
- American Society for Microbiology
- American Society of Hematology
- American Society of Human Genetics
Editorial Positions and Referee Activities
- 2022 – Review Editor, Frontier in Cellular and Infection Microbiology (Frontiers)
- 2022 – Reviewer, Frontier Medicine (Frontiers)
- 2022 – Reviewer, mBio (American Society for Microbiology)
- 2020 – Reviewer, Nature Biotechnology (Springer Nature)
- 2020 – Reviewer, AIDS Research and Human Retroviruses (Marry Ann Liebert)
- 2019 – Reviewer, Virology Journal (Springer)
- 2019 – Grant Reviewer, NIH Novel Genomic Technology Development Special Emphasis Panel (ZRG1 GGG-T [50])
- 2017 – Grant reviewer, Amfar ARCHE-GT grant (the foundation for AIDS Research)
- 2016 – Reviewer, Archives of Virology (Springer)
- 2008 – Reviewer, Virology (Elservier)
- 2007 – Reviewer, Journal of Virology (American Society for Microbiology)
Awards and Honors
2017 – 2020
- American Society of Hematology Scholar Award
ASH scholar award is “One of ASH’s most prestigious award programs” providing $150,000 for junior faculty over three-year period.
2012 – 2020
- NIH K99/R00 Pathway to Independence Award (1K99HL116234)
2014
- Young Investigator Award, the International Antiviral Society-USA, Conference on Retroviruses and Opportunistic Infections,
2013
- Palm Springs Symposium on HIV/AIDS - Travel Award
2011
- California Institute of Regenerative Medicine Scholarship Award
2011
- Stem Cell Engineering & Cell Therapy - Travel Award. Cold Spring Harbor Laboratory.
2008 – 2009
- UCLA AIDS Institute Postdoctoral Fellowship
2006 – 2007
- UCLA Dissertation Year Fellowship Award
2004
- American Society of Microbiology - Travel Award.
Publications
Research Articles
- Yu H, Golconda S, Lee GE, Xue D, Dominguez-Huerta G, Wainaina JM, Bolduc B, Murthy S, Kim S, Faith S, Liu SL, Lee J, Oglesbee M, Sullivan MB, Kim S. "CLAE: A High-Fidelity Nanopore Sequencing Strategy for Read-Level Viral Variant Detection and Environmental RNA Virus Discovery." Adv Sci (Weinh). 2025 Sep 11:305978. doi:10.1002/advs.202505978. Online ahead of print. PMID: 40936099 *Senior corresponding author
- King EM, Midkiff A, McClain K, Kim S, Panfil AR. "YTHDF1 and YTHDC1 m6A reader proteins regulate HTLV-1 tax and hbz activity." J Virol. 2025 Mar 18;99(3):e0206324. doi:10.1128/jvi.02063-24. Epub 2025 Feb 4.
- PMID: 39902961 Baek A, Lee GE, Golconda S, Rayhan A, Manganaris A, Chen S, Tirumuru N, Yu H, Kim S, Kimmel C, Zablocki O, Sullivan M, Addepalli B, Wu L, Kim S*, “Single-molecule epitranscriptomic analysis of full-length HIV-1 RNAs reveals functional roles of site-specific m6As”, Nature Microbiology, 2024 May;9(5):1340-1355. doi:10.1038/s41564-024-01638-5. Epub 2024 Apr 11. PMID: 38605174 Free PMC article. *Senior corresponding author
- Baek A, Rayhan A, Lee GE, Golconda S, Yu H, Kim S, Limbach PA, Addepalli B, Kim S*. Mapping m6A Sites on HIV-1 RNA Using Oligonucleotide LC-MS/MS. Methods Protoc. 2024 Jan 10;7(1). PubMed PMID: 38251200; PubMed Central PMCID: PMC10801558. doi: 10.3390/mps7010007. *Senior corresponding author
- Wang L, Suryawanshi GW, Kim S, Guan X, Bonifacino AC, Metzger ME, Donahue RE, Kim S, Chen ISY. CD3-immunotoxin mediated depletion of T cells in lymphoid tissues of rhesus macaques. Heliyon. 2023 Sep; 9(9): e19435 Sep. PubMed PMID: 37810095; PMC10558572. . doi: 10.1016/j.heliyon.2023.e19435. eCollection 2023
- Kim S, Shukla RK, Yu H, Baek A, Cressman SG, Golconda S, Lee GE, Choi H, Reneau JC, Wang Z, Huang CA, Liyanage NPM, Kim S*. CD3e-immunotoxin spares CD62L(lo) Tregs and reshapes organ-specific T-cell composition by preferentially depleting CD3e(hi) T cells. Front Immunol. 2022; 13:1011190. PubMed PMID: 36389741; PubMed Central PMCID: PMC9643874. doi: 10.3389/fimmu.2022.1011190. eCollection 2022. *Senior corresponding author
- Phillips S, Baek A, Kim S, Chen S, Wu L. Protocol for the generation of HIV-1 genomic RNA with altered levels of N6-methyladenosine. STAR Protoc. 2022 Sep 16;3(3):101616 Sep 16. PubMed PMID: 35990737; PubMed Central PMCID: PMC9389413. . doi: 10.1016/j.xpro.2022.101616. eCollection 2022
- Chiang CL, Hu EY, Chang L, Labanowska J, Zapolnik K, Mo X, Shi J, Doong TJ, Lozanski A, Yan PS, Bundschuh R, Walker LA, Gallego-Perez D, Lu W, Long M, Kim S, Heerema NA, Lozanski G, Woyach JA, Byrd JC, Lee LJ, Muthusamy N. Leukemia-initiating HSCs in chronic lymphocytic leukemia reveal clonal leukemogenesis and differential drug sensitivity. Cell Rep. 2022 Jul 19;40(3):111115. doi: 10.1016/j.celrep.2022.111115. PubMed PMID: 35858552.
- Kim S, Shukla RK, Kim E, Cressman SG, Yu H, Baek A, Choi H, Kim A, Sharma A, Wang Z, Huang CA, Reneau JC, Boyaka PN, Liyanage NPM, Kim S*. Comparison of CD3e Antibody and CD3e-sZAP Immunotoxin Treatment in Mice Identifies sZAP as the Main Driver of Vascular Leakage. Biomedicines. 2022 May 24;10(6). PubMed PMID: 35740248; PubMed Central PMCID: PMC9220018. doi: 10.3390/biomedicines10061221. *Senior corresponding author
- Kim DH, Choi YM, Lee J, Shin S, Kim S, Suh Y, Lee K. Differential Expression of MSTN Isoforms in Muscle between Broiler and Layer Chickens. Animals (Basel). 2022 Feb 22;12(5). PubMed PMID: 35268106; PubMed Central PMCID: PMC8908836. doi: 10.3390/ani12050539.
- Suryawanshi GW, Arokium H, Kim S, Khamaikawin W, Lin S, Shimizu S, Chupradit K, Lee Y, Xie Y, Guan X, Suryawanshi V, Presson AP, An DS, Chen ISY. Longitudinal clonal tracking in humanized mice reveals sustained polyclonal repopulation of gene-modified human-HSPC despite vector integration bias. Stem Cell Res Ther. 2021 Oct 7;12(1):528. doi: 10.1186/s13287-021-02601-5. PubMed PMID: 34620229; PubMed Central PMCID: PMC8499514.
- Kim DH, Lee J, Kim S, Lillehoj HS, Lee K. Hypertrophy of Adipose Tissues in Quail Embryos by in ovo Injection of All-Trans Retinoic Acid. Front Physiol. 2021;12:681562. doi: 10.3389/fphys.2021.681562. eCollection 2021. PubMed PMID: 34093239; PubMed Central PMCID: PMC8176229.
- Gregory AC, Zablocki O, Zayed AA, Howell A, Bolduc B, Sullivan MB. The Gut Virome Database Reveals Age-Dependent Patterns of Virome Diversity in the Human Gut. Cell Host Microbe. 2020 Nov 11;28(5):724-740.e8. doi: 10.1016/j.chom.2020.08.003. Epub 2020 Aug 24. PubMed PMID: 32841606; PubMed Central PMCID: PMC7443397.
- Suryawanshi GW, Khamaikawin W, Wen J, Shimizu S, Arokium H, Xie Y, Wang E, Kim S, Choi H, Zhang C, Yu H, Presson AP, Kim N, An DS, Chen ISY, Kim S*. The clonal repopulation of HSPC gene modified with anti-HIV-1 RNAi is not affected by preexisting HIV-1 infection. Science Advances (AAAS). 2020 Jul;6(30):eaay9206. Jul. PubMed PMID: 32766447; PubMed Central PMCID: PMC7385479. doi: 10.1126/sciadv.aay9206. eCollection 2020 *Senior corresponding author
- Xu S, Kim S, Chen ISY, Chou T. Modeling large fluctuations of thousands of clones during hematopoiesis: The role of stem cell self-renewal and bursty progenitor dynamics in rhesus macaque. PLoS Comput Biol. 2018 Oct;14(10): e1006489. doi: 10.1371/journal.pcbi.1006489. eCollection 2018 Oct. PubMed PMID: 30335762; PubMed Central PMCID: PMC6218102.
- Khamaikawin W, Shimizu S, Kamata M, Cortado R, Jung Y, Lam J, Wen J, Kim P, Xie Y, Kim S, Arokium H, Presson AP, Chen ISY, An DS. Modeling Anti-HIV-1 HSPC-Based Gene Therapy in Humanized Mice Previously Infected with HIV-1. Molecular therapy. Methods & clinical development. 2018; 9:23-32. PubMed [journal] PMID: 29322065, PMCID: PMC5751878
- Suryawanshi GW, Xu S, Xie Y, Chou T, Kim N, Chen ISY, Kim S*. Bidirectional Retroviral Integration Site PCR Methodology and Quantitative Data Analysis Workflow. Journal of visualized experiments : JoVE. 2017; (124). PubMed [journal] PMID: 28654067, PMCID: PMC5608418 *Senior corresponding author
- Goyal S*, Kim S*, Chen IS, Chou T. Mechanisms of blood homeostasis: lineage tracking and a neutral model of cell populations in rhesus macaques. BMC Biol. 2015 Oct 20;13:85. doi: 10.1186/s12915-015-0191-8. PubMed PMID: 26486451; PubMed Central PMCID: PMC4615871.
- Kim S, Kim N, Presson AP, Metzger ME, Bonifacino AC, Sehl M, Chow SA, Crooks GM, Dunbar CE, An DS, Donahue RE, Chen IS. Dynamics of HSPC repopulation in nonhuman primates revealed by a decade-long clonal-tracking study. Cell Stem Cell. 2014 Apr 3;14(4):473-85. doi: 10.1016/j.stem.2013.12.012. PubMed PMID: 24702996; PubMed Central PMCID: PMC4009343.
- Arokium H, Kamata M*, Kim S*, Kim N, Liang M, Presson AP, Chen IS. Deep sequencing reveals low incidence of endogenous LINE-1 retrotransposition in human induced pluripotent stem cells. PLoS One. 2014 Oct 7;9(10):e108682. doi: 10.1371/journal.pone.0108682. eCollection 2014. PubMed PMID: 25289675; PubMed Central PMCID: PMC4188539.
- Kim S, Kim YC, Qi H, Su K, Morrison SL, Chow SA. Efficient identification of human immunodeficiency virus type 1 mutants resistant to a protease inhibitor by using a random mutant library. Antimicrob Agents Chemother. 2011 Nov;55(11):5090-8. doi: 10.1128/AAC.00687-11. Epub 2011 Aug 29. PubMed PMID: 21876045; PubMed Central PMCID: PMC3195011.
- Presson AP, Kim N, Xiaofei Y, Chen IS, Kim S. Methodology and software to detect viral integration site hot-spots. BMC Bioinformatics. 2011 Sep 14;12:367. doi: 10.1186/1471-2105-12-367. PubMed PMID: 21914224; PubMed Central PMCID: PMC3203353.
- Kim S, Kim N, Presson AP, An DS, Mao SH, Bonifacino AC, Donahue RE, Chow SA, Chen IS. High-throughput, sensitive quantification of repopulating hematopoietic stem cell clones. J Virol. 2010 Nov;84(22):11771-80. doi: 10.1128/JVI.01355-10. Epub 2010 Sep 15. PubMed PMID: 20844053; PubMed Central PMCID: PMC2977897.
- Kim S, Rusmevichientong A, Dong B, Remenyi R, Silverman RH, Chow SA. Fidelity of target site duplication and sequence preference during integration of xenotropic murine leukemia virus-related virus. PLoS One. 2010 Apr 20;5(4):e10255. doi: 10.1371/journal.pone.0010255. PubMed PMID: 20421928; PubMed Central PMCID: PMC2857682.
- Shimizu S, Kamata M, Kittipongdaja P, Chen KN, Kim S, Pang S, Boyer J, Qin FX, An DS, Chen IS. Characterization of a potent non-cytotoxic shRNA directed to the HIV-1 co-receptor CCR5. Genet Vaccines Ther. 2009 Jun 10;7:8. doi: 10.1186/1479-0556-7-8. PubMed PMID: 19515239; PubMed Central PMCID: PMC2701936.
- Kim S*, Vatakis DN*, Kim N, Chow SA, Zack JA. Human immunodeficiency virus integration efficiency and site selection in quiescent CD4+ T cells. J Virol. 2009 Jun;83(12):6222-33. doi: 10.1128/JVI.00356-09. Epub 2009 Apr 15. PubMed PMID: 19369341; PubMed Central PMCID: PMC2687367.
- Kim S, Kim N, Dong B, Boren D, Lee SA, Das Gupta J, Gaughan C, Klein EA, Lee C, Silverman RH, Chow SA. Integration site preference of xenotropic murine leukemia virus-related virus, a new human retrovirus associated with prostate cancer. J Virol. 2008 Oct;82(20):9964-77. doi: 10.1128/JVI.01299-08. Epub 2008 Aug 6. PubMed PMID: 18684813; PubMed Central PMCID: PMC2566297.
- Dong B, Kim S, Hong S, Das Gupta J, Malathi K, Klein EA, Ganem D, Derisi JL, Chow SA, Silverman RH. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc Natl Acad Sci U S A. 2007 Jan 30;104(5):1655-60. Epub 2007 Jan 18. PubMed PMID: 17234809; PubMed Central PMCID: PMC1776164.
- Kim S, Kim Y, Liang T, Sinsheimer JS, Chow SA. A high-throughput method for cloning and sequencing human immunodeficiency virus type 1 integration sites. J Virol. 2006 Nov;80(22):11313-21. Epub 2006 Sep 13. PubMed PMID: 16971446; PubMed Central PMCID: PMC1642152.
- Kim Y, Kim S, Earnest TN, Hol WG. Precursor structure of cephalosporin acylase. Insights into autoproteolytic activation in a new N-terminal hydrolase family. J Biol Chem. 2002 Jan 25;277(4):2823-9. Epub 2001 Nov 8. PubMed PMID: 11706000.
- Kim S, Kim Y. Active site residues of cephalosporin acylase are critical not only for enzymatic catalysis but also for post-translational modification. J Biol Chem. 2001 Dec 21;276(51):48376-81. Epub 2001 Oct 16. PubMed PMID: 11604409.